AS Database
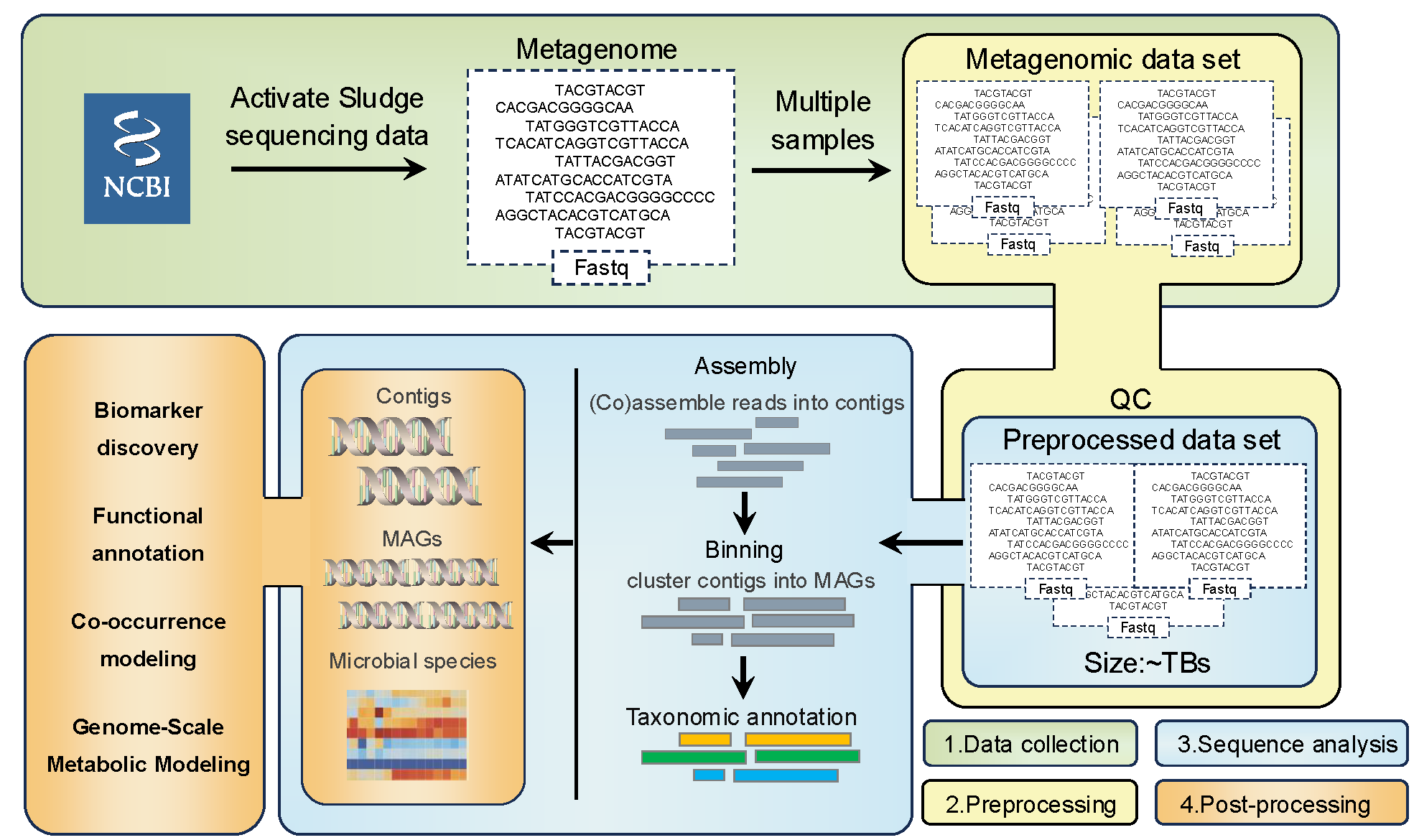
To investigate microbial diversity within activated sludge and assess its global distribution, we assembled a comprehensive metagenomic dataset derived from 469 Bioproject IDs obtained from the National Center for Biotechnology Information (NCBI). This dataset comprises 287 activated sludge samples from 13 countries, amounting to 14.68 terabytes of raw sequence data. Each sequence undergoes a rigorous processing workflow, including quality control, trimming, assembly, binning, taxonomic classification, and estimation of species relative abundance. Ultimately, our database yields 2,326 medium- to high-quality metagenome-assembled genomes (MAGs), facilitating an in-depth taxonomic classification of bacterial species. By providing a detailed catalog of microbial species, their relative abundances, and global distribution patterns within activated sludge, this dataset establishes a valuable foundation for future research on microbial communities in this essential wastewater treatment system.
Contents
Global Sample Site Distribution: Geographic documentation of sampling locations, capturing activated sludge samples from diverse countries and regions.
Continuous Microbial Sequence Fragments: Includes contigs representing continuous sequence fragments generated during data processing, providing foundational data for further analysis.
High-Quality Microbial Genome Sequences: Contains high-quality metagenome-assembled genomes (MAGs) representing microbial genomes specific to activated sludge communities.
Capabilities
Comprehensive Species Cataloging: An extensive catalog of microbial species within activated sludge, with detailed taxonomic classification across hierarchical ranks (e.g., domain, kingdom, phylum, genus, species) encompassing bacteria, archaea, and other microorganisms.
Diversity and Abundance Analysis: Facilitates large-scale analysis of microbial diversity and species abundance within sludge samples on a global scale.
Functional Annotation of MAGs: Supports functional annotation of MAGs using tools such as Prokka and KEGG, enabling detailed functional analysis of genes associated with key traits, including metabolism and resistance, thereby providing insights into microbial functional roles.
Genome-Scale Metabolic Modeling (GEM): High-quality genome data supports the construction of genome-scale metabolic models, enabling the simulation and analysis of metabolic networks within activated sludge microbial communities.